HBC
Quantitative Assay: Protein Microarray

Protein microarrays are microscope slides, printed with thousands of tiny spots in defined positions. They allow for rapid, economical and automated handling of highly sensitive materials, and they were developed due to the limitations of using DNA microarrays for determining gene expression levels in proteomics, which is the large-scale study of proteins. They replaced traditional proteomics techniques, which were time-consuming, labour intensive and not very well suited for the analysis of low abundant proteins.
1983 saw the first introduction to protein microarrays and their concept and methodology in a scientific publication (https://www.sciencedirect.com/science/article/abs/pii/0022175983903186?via%3Dihu) and in a series of patents. Since the technology for protein microarrays was inspired by technology developed for DNA and RNA microarrays, it was fairly straight-forward to develop the high-throughput technology needed.
Process of contact printing/spotting of protein microarrays:
Large numbers of proteins or their ligands are robotically placed onto a coated, solid surface/support in a pre-defined pattern. This is the most common type of protein microarray.
Process of non-contact, piezoelectric spotting of protein microarrays:
The printhead moves across the array and uses electric stimulation at each spot to deliver the protein solution onto the surface as tiny droplets.
Using this technology, proteins are arrayed onto a solid surface, e.g. glass slides, membranes, or at the bottom of 96-well microtiter plates. The surface provides a support onto which recombinant proteins or allergens can be immobilised. A serum is incubated with these proteins under consistent conditions, and the antibodies present in the serum bind to their target (antigen) if present on the surface. Colorimetric or fluorescent detectors are then used to detect bound antibodies by the use of a labelled secondary antibody in between numerous rounds of washing.
As a result of the miniaturisation and high degree of multiplexing made possible by using such microarrays instead of more traditional methods, such as Western Blotting, a significant reduction in the volume of precious sample (e.g. blood), as well as expensive reagents (secondary antibody), can be achieved.
Typical examples of the types of capture molecules arrayed on a solid surface are:
- antibodies
- antigens
- Peptides as potential epitopes
- aptamers
- affibodies
- full length proteins
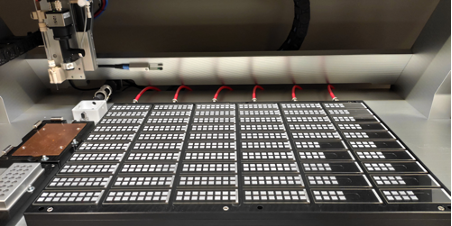
To immobilise the proteins on the surface, this is coated with a thin layer of nitrocellulose, or a PVDF membrane is used instead, or attached to the surface. High density arrays are produced by depositing the different spots in close proximity to one another using our piezo dispensing technology.
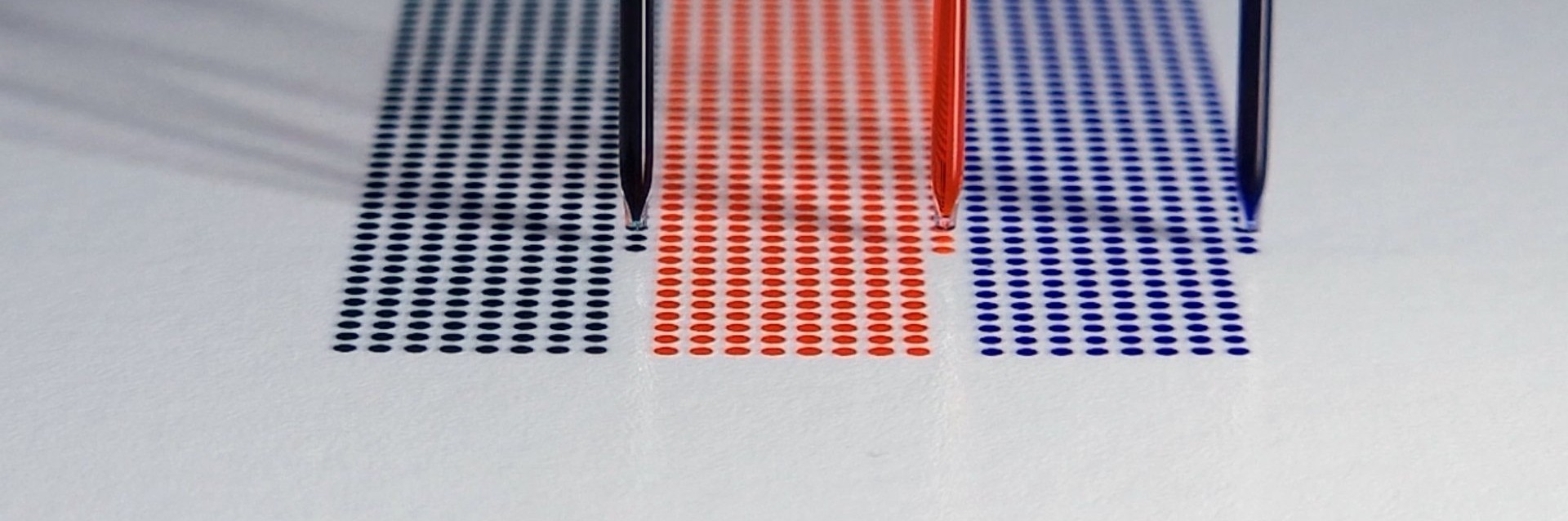
Obviously, an exceptionally precise deposition system is needed that can handle volume transfer in the low to mid-picolitre range with high accuracy. The protein spot-to-spot distance (centre to centre) should be smaller than 500 µm and ideally fall below 200 µm.
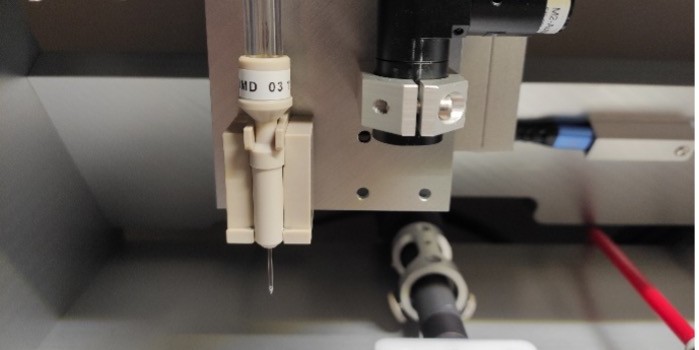
The iONE-600 dispensing-instrument from M2-Automation offers the accuracy, speed and robustness needed for the production of high-quality protein microarrays. As droplet deposition is achieved without contact with the surface, the latter is excluded as cause for contamination and sample carry-over.
Preparation elements of a microarray
A completed microarray
The development and quantification processes are complete. The distance between spots is about 500 um, and each spot is about 1 nL (3 droplets of about 330 pL each).